13. Good practice in relational database design
When building databases we should be careful to reduce the amount of redundant data – in an ideal world, the tables of our database should not contain any repeated data, ensuring that data points are not repeated in our database.
This is called data normalisation. It is the process of correctly disaggregating data sets, such that each data point is atomic – represented only a single time and efficiently represented by the relational model.
The principal benefits of this update your database will be logically arranged such that the data has no repetition or redundancy. This in turn, allows it to be efficiently stored and queried in most modern database systems.
The objectives of normalisation beyond 1NF (first normal form) were stated as follows by Codd:
- To free the collection of relations from undesirable insertion, update and deletion dependencies.
- To reduce the need for restructuring the collection of relations, as new types of data are introduced, and thus increase the life span of application programs.
- To make the relational model more informative to users.
- To make the collection of relations neutral to the query statistics, where these statistics are liable to change as time goes by.
— E.F. Codd, “Further Normalisation of the Data Base Relational Model”
1NF (first normal form) – this requires that our data is indexed with a unique primary key, that there are no repeating unit groups, and that our data is atomic.
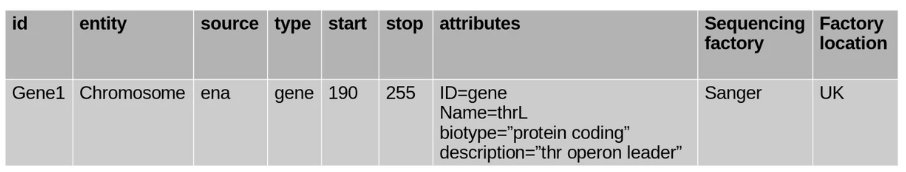
Consider this table:
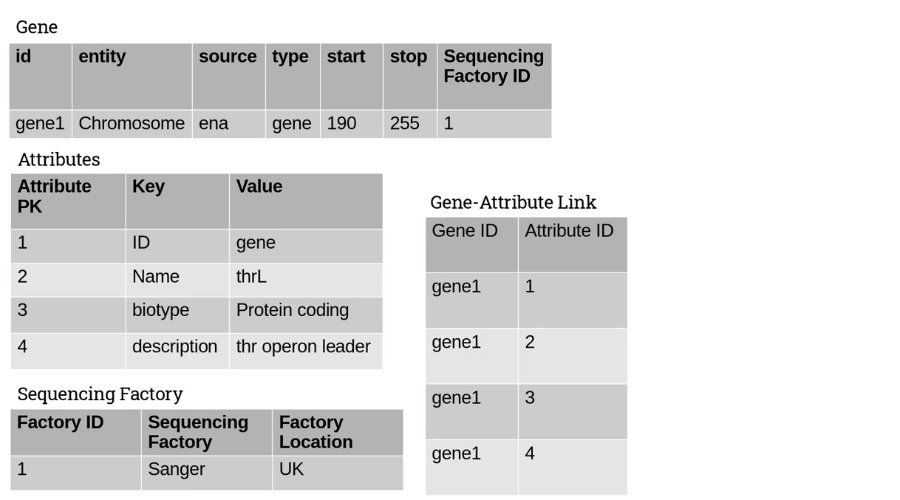
It would make sense for the attributes to be taken out and given their own table. Also, the factory could come up again and again, so it would be better to create a factory table and link it back with a foreign key.
Also – each gene could have some or all of the attributes, so to prevent further redundancy we can create a new table linking the genes and the attributes:
This now brings us into 1NF – data is indexed with a unique primary key, there are no repeating unit groups, and that our data is atomic.
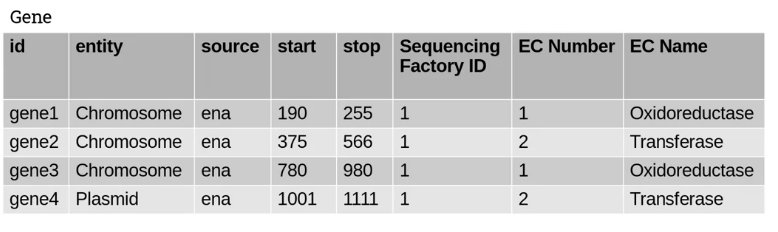
2NF (second normal form) requires that we remove any columns which may predict each other. Consider another problem, with this new table:
You can see that two columns – EC Number and EC Name – seem to depend on each other. We can predict the content of EC Name from EC Number, and vice versa.
We should be able to extract these columns into a new table and use a foreign key to link them, as follows:
This ensures our dataset is now in 3NF (third normal form).
We end up with a series of tables that remove any data redundancy:
Here is how we can set these tables up referencing each other as explained above:
CREATE DATABASE gene_test; CREATE TABLE ec(pk SERIAL PRIMARY KEY, EC_name VARCHAR(256)); CREATE TABLE sequencing(pk SERIAL PRIMARY KEY, sequencing_factory VARCHAR(256), factory_location VARCHAR(256)); CREATE TABLE genes (pk SERIAL PRIMARY KEY, gene_id VARCHAR(256) NOT NULL, entity VARCHAR(256), source VARCHAR(256), start INT, stop INT, sequencing_pk INT, ec_pk INT, FOREIGN KEY (sequencing_pk) REFERENCES sequencing(pk), FOREIGN KEY (ec_pk) REFERENCES ec(pk)); CREATE TABLE products (genes_pk INT, type VARCHAR(256), product VARCHAR(256), FOREIGN KEY (genes_pk) REFERENCES genes(pk)); CREATE TABLE attributes (pk SERIAL PRIMARY KEY, key VARCHAR(256), value VARCHAR(256)); CREATE TABLE gene_attribute_link(genes_pk INT, attributes_pk INT, FOREIGN KEY (genes_pk) REFERENCES genes(pk), FOREIGN KEY (attributes_pk) REFERENCES attributes(pk));
Wednesday 27 October 2021, 592 views
Next post: 14. Inserts and queries in SQL Previous post: 12. Limitations to database modelling
Advanced Web Development index
- 38. Writing API tests
- 37. Testing in Django
- 36. Class-based views in the Django REST framework
- 35. Building a RESTful web service in Django
- 34. Introduction to CRUD, REST and APIs
- 33. Refactoring with generic views in Django
- 32. Django validators
- 31. Django forms (2) – using the ModelForm class
- 30. Django forms (1)
- 29. JavaScript basics
- 28. Adding CSS to the template
- 27. Django templating
- 26. Deleting and updating records
- 25. Joins, filters and chaining commands
- 24. Using the ORM in views.py
- 23. Adding to the database by writing a script
- 22. Adding to the database with Django Admin
- 21. Migrations
- 20. ORM – work through example
- 19. An introduction to the Object-Relational Mapper
- 18. Altering the database
- 17. SQL functions and summaries
- 16. SQL Query performance
- 15. Queries and table joins in SQL
- 14. Inserts and queries in SQL
- 13. Good practice in relational database design
- 12. Limitations to database modelling
- 11. Building a database using SQL
- 10. Introduction to PostgreSQL
- 9. How to start writing a new application in Django
- 8. Building a lightweight project
- 7. Django URLs
- 6. Django templates
- 5. Django models
- 4. Django views
- 3. Creating a new hello app
- 2. Creating a new virtual environment
- 1. Setting up Django



Leave a Reply